This research along with its peer reviewed paper and app development was possible thanks to NIH BRAIN Initiative Grant # R01 EB028157
In our new paper titled “Fibration symmetries and cluster synchronization in the Caenorhabditis elegans connectome” accepted on January 26, 2024, on PLOS ONE (soon to be published), we explore the structural properties that give rise to synchronization patterns of neurons in this magnificent creature’s motor-neuron system. Additionally, we introduce a new measure for quantifying synchronization.
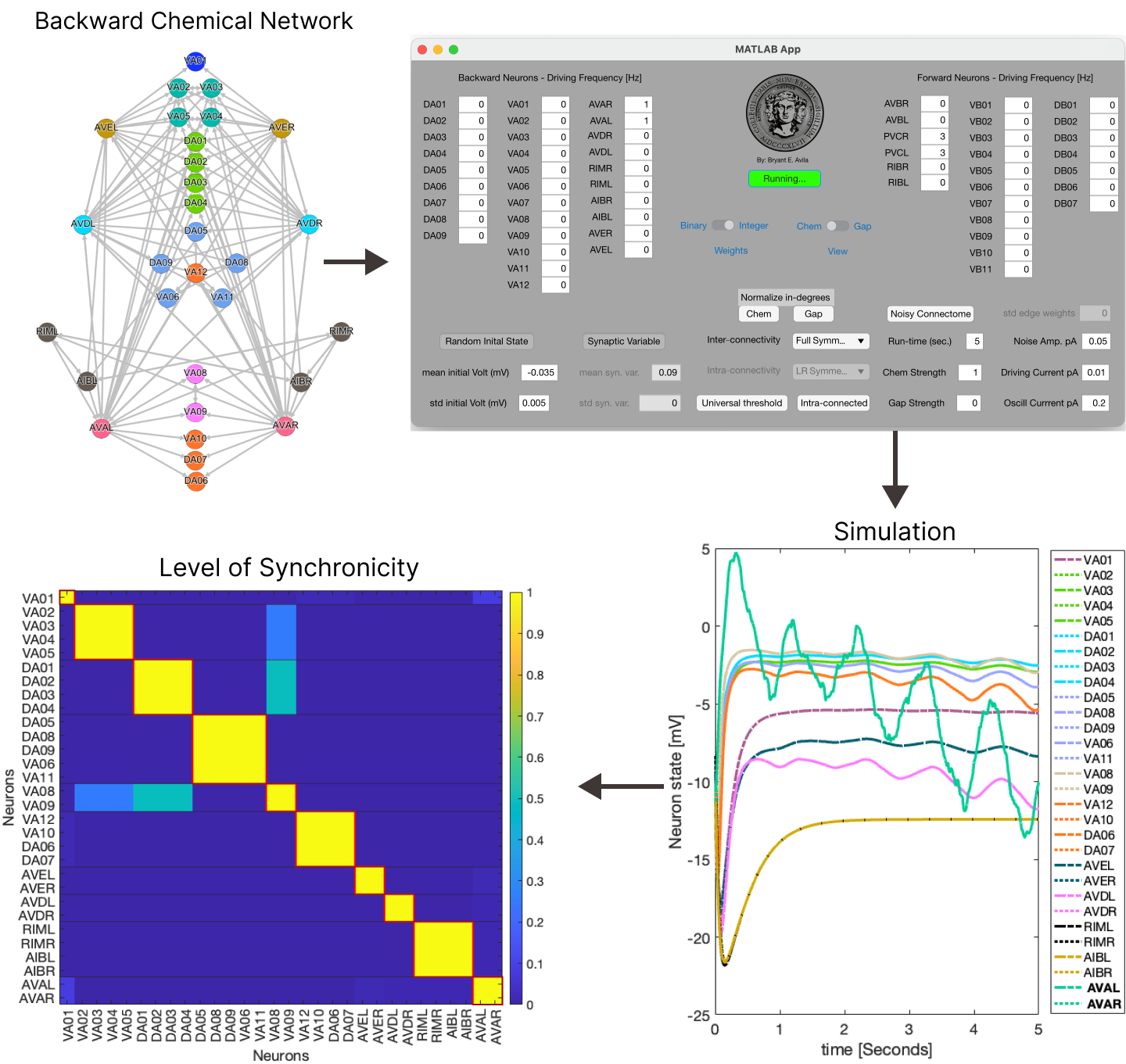
Along with this paper we make available a friendly to use GUI app built on Matlab which the user can use to explore how different portions of the C. elegans locomotive neural system synchronize (or not) depending on the multiple factors. This gives the public a plethora of knobs and cranks to better understand a fundamental property of network symmetries (fibrations) and neuron interactions!
The user can specify the type of connectomes used such as (electron micrograph connectivity produced by) Varshney, its left-right symmetrized and fully fiber symmetrized versions for the forward and backward locomotive systems. Along with this one can specify the types of symmetrizations for the inter-connectivity between the forward and backward systems, where interactions between these are considered inhibitory while connection within each system (intra-connectivity) are considered excitatory.
These simulations are enabled by using ordinary differential equation models that emulate neuron interactions. These equations can be seen below and are compromised by the two main means of neuronal interactions: gap junctions and chemical synapses.
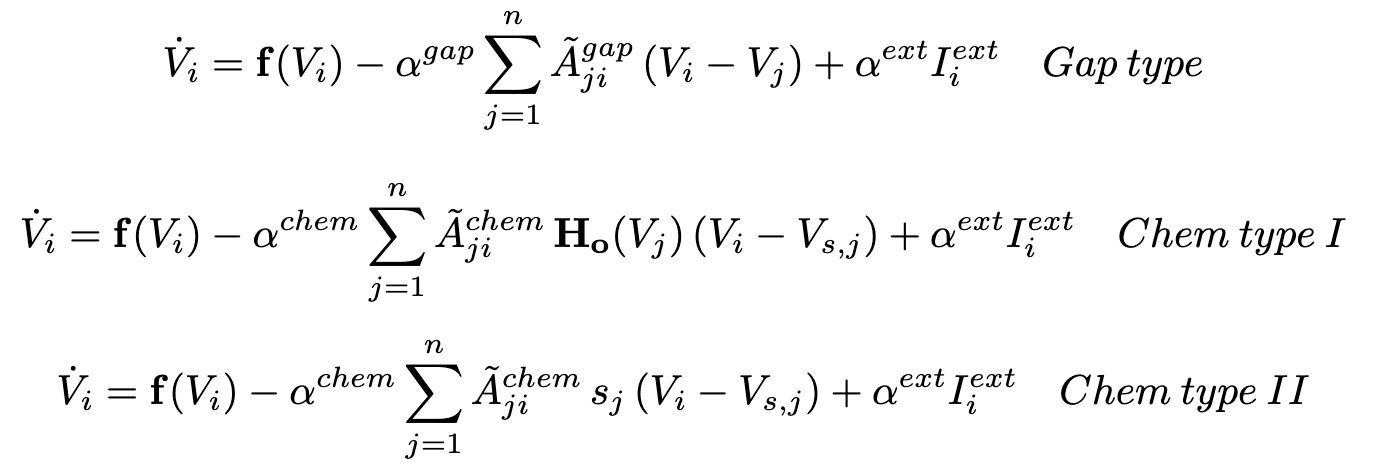
The strength of the gap junctions and chemical synapses interactions can be specified and allow a network to only have one of either of these interactions or any combination of strength between these two. Additionally, it is possible to specify the type of weights used for the connectivity (binary or integer) and the ability to randomly alter these weights, normalize the in-degree of each neuron for both types of interactions, the initial distribution of voltages and synaptic variables, type of model used for the chemical synapsis interactions and more controls. Finally, one is able to specify which neurons receive a static input current, the amplitude and frequency of an undulatory input and a Gaussian random walk input.
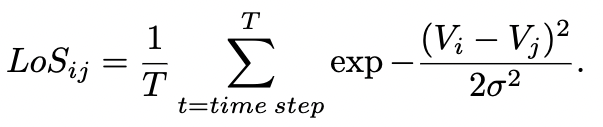
After the simulation has concluded the app calculates the level of synchronicity (LoS) as per the equation above for the pair of neurons for each of the two networks. This is an important measure introduced into the field as it captures the synchronization between two signals as stimulate by fibration theory. Other measurements are also accessible (Phase Locking Value, Phase Difference and Standard Deviation Difference).
This research along with its peer reviewed paper and app development was possible thanks to NIH BRAIN Initiative Grant # R01 EB028157
